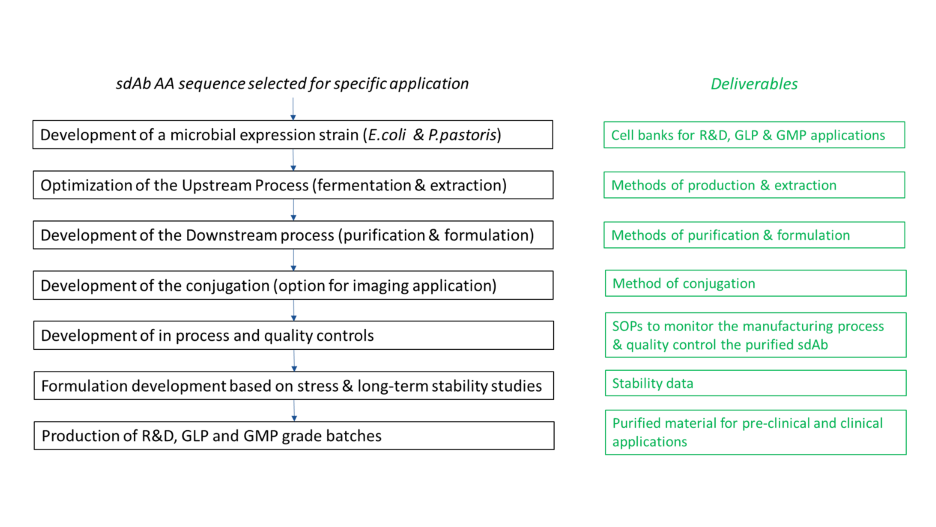
Summary
This application note gives an extensive process development approach to establish an optimized manufacturing protocol for the production of single-domain antibodies (sdAb) for therapeutic purposes. This includes microbial expression strain developement, upstream and downstream process optimization, and formulation development, leading to High-quality (HQ ) and good manufacturing practice (GMP) grade production.
Introduction
The camelid-derived single– chain antibody (sdAb) is a unique antibody fragment with a heavy-chain variable domain. This small molecular weight (12-15 kDa) antibody, exhibits similar antigen-binding affinity to conventional antibody, and has different advantageous features for various applications. Indeed, their high solubility and stability, their accessibility to a larger number of epitopes, as well as their high manufacturability make them candidates of choice for different applications such as oncology, immune therapies, infection diseases, imaging and therapeutics delivery to the central nervous system (CNS) due to their ability to cross the blood-brain barrier.
Xpress Biologics set up a complete platform specifically adapted to the production of sdAb for pre-clinical to clinical applications (Fig.1). For this, two microbial expression systems are proposed for the expression of sdAb: Escherichia coli and Pichia pastoris.
*GLP : Good Laboratory Practice
Development of a microbial expression strain (E.coli)
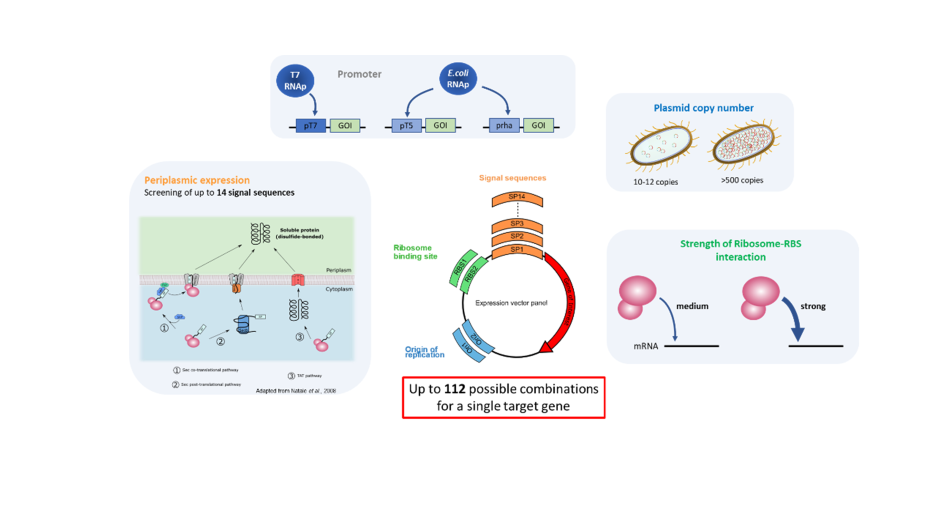
The molecular biology toolbox set up by Xpress Biologics to rapidly select an efficient Escherichia coli expression system is also applicable for the sdAb. The key parameter to ensure an efficient expression in the Escherichia coli (E.coli) periplasmic compartment lies in the synchronization between the synthesis and secretion machineries. Different attributes impacting the synthesis and secretion efficiency, such as gene copy number, transcription efficiency and translation strength, as well as different secretion pathways, are therefore tested (Fig.2).
Selection of the most efficient expression system follows a 4-weeks process (Fig.3):
After the design and the sequence optimization of the sdAb DNA sequence for E. coli expression and the gene synthesis, the sequence is cloned in up to 14 different vectors exhibiting different signal peptides, allowing the evaluation of different secretion pathways: the Sec translocation pathway– specific to the secretion of unfolded proteins and folding in the periplasm, the TAT pathway and the SRP pathway which are better adapted for the secretion of quick folding proteins.
The sdAb expression the 14 clones at small- scale culture (1 ml culture).Three different induction conditions (low, medium, and high) are performed for each clone to evaluate, for each signal peptide, the expression level’s effect on secretion efficiency. The periplasmic content is recovered, and the sdAb production yield is evaluated by SDS-PAGE analysis. The 2 signal sequences leading to the highest periplasmic expression are selected to assess the efficiency of the T7 promoter, different ORI, and RBS.
The effect of the strong T7 promoter (IPTG inducible) in combination with 2 Origins of Replication (for low and high plasmid copy number) and 2 different RBS (medium and high binding) on the sdAb periplasmic expression using the 2 selected signal sequences is therefore evaluated (8 vectors in total). The 2 DNA sequences coding for each signal peptide-sdAb open-reading-frames are amplified by PCR and cloned into the 4 vectors before transformation in E.coli. Assessment of the sdAb production efficiency between the 8 different clones is performed at small-scale (1ml culture), the sdAb production yield being evaluated by SDS-PAGE analysis. A glycerol cell bank is produced for the best 4 expression systems, and their expression yield is compared in conditions mimicking industrial fermentation process.
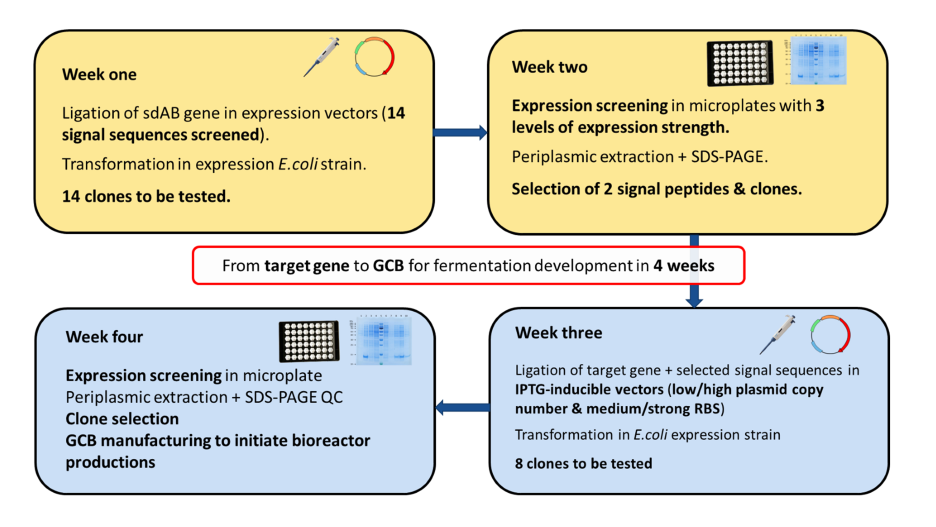
Fig.3 – Four-weeks process for the selection of an efficient E.coli expression system
Optimization of the Upstream Process
Our platform of optimization of fermentation conditions allowing the best sdAb expression is based on an experimental plan performed in 5 L-scale fermenters. The fermentation development platform includes a set of four identical stainless-steel fermenters (Biostat® C+, Sartorius) with a working volume of 5 L, which are representative to the fermenters used in the industrial sector, as well as one 50 L-scale fermenter (Biostat® D-DCU, Sartorius) for scaling-up studies.
The standard protocol developed by Xpress Biologics corresponds to a high cell– density fed-batch fermentation. The objective of the optimization plan is to test the impact of different parameters upon the expression yield and the protein integrity (examples: biomass at induction, temperature & pH, type of substrates, growth rate, and duration of induction). All fermentation runs are monitored using the following parameters: dissolved oxygen, pressure, airflow, agitation speed, temperature, pH, acid / base consumption, and antifoam addition.
The selection of the best fermentation conditions is based on different microbiological tests :
- CFU, OD600 & wet cell weight
- Kinetic of expression after induction
- Final expression yield
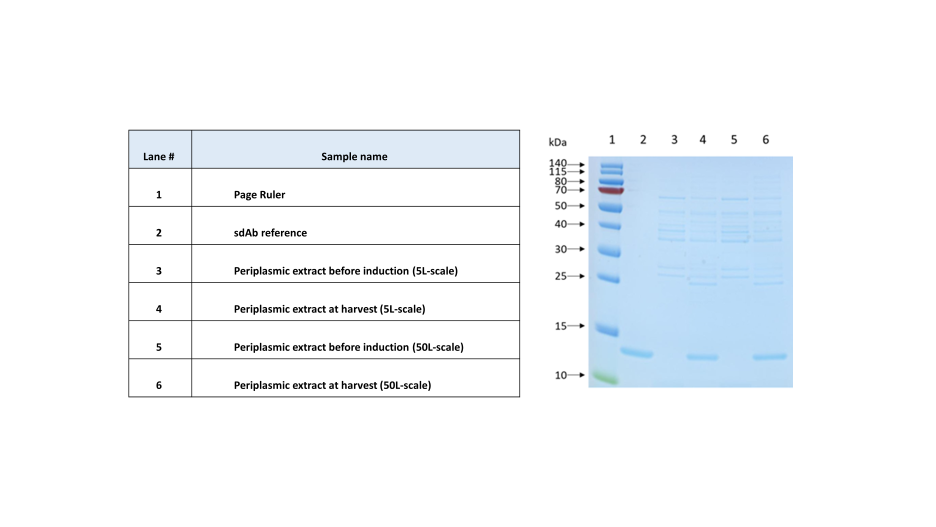
The expression level of a sdAb recorded using the conditions and parameters optimized at a 5L fermentation scale has been compared to the one obtained at a 50L scale (Fig.4). Based on a densitometry analysis, the expression levels are found to be very close: 1.82 and 1.71 g/L, respectively. It is also important to note that the host cell proteins profile (HCP) is also very similar between both scales, demonstrating once again the scalability of the developed fermentation process.
Development of the downstream process
The strategy to purify a sdAb is based on the development of 1 capture chromatography followed by one polishing chromatography and one ultrafiltration step for the formulation of the purified protein (Fig.5).
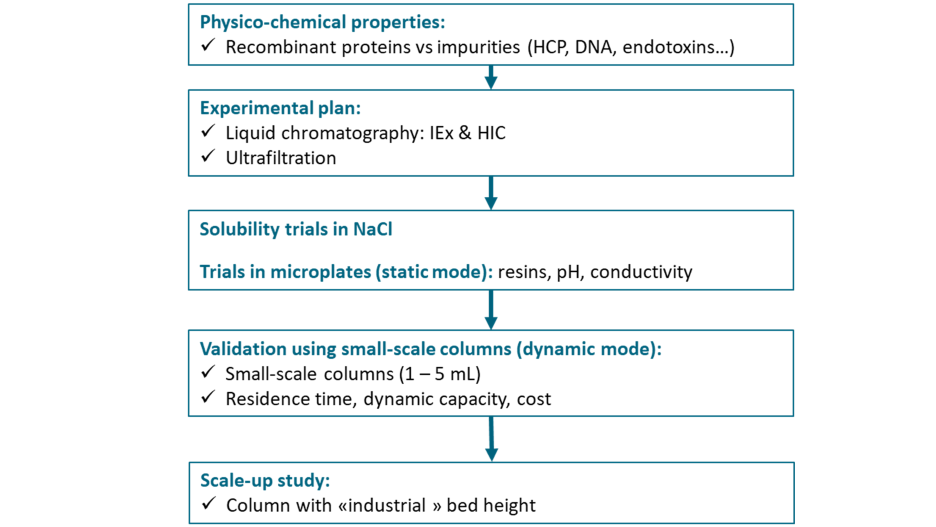
Fig.5 – Strategy to develop a purification process for a sdAb
The choice between different types of chromatographic supports (anion and/or cation exchange and hydrophobic interaction sorbents) for the 1st and 2nd chromatographic steps is based on the physico–chemical properties of the protein (hydrophobic profile, pI). A preliminary solubility trial is performed to define the operating ranges (i.e., pH and salt concentrations) that can be applied during the purification development. The conditions of loading, washing, and elution (protein load, buffer type, pH, and salt concentration), for each matrix, have been tested based on a Design of Experiment (DoE) a mythology that uses controlled tests to evaluate the individual and interactive effects of various factors on an outcome of a process, approach performed using 96-well plates (static mode). The purity improvement, the host cell proteins (HCPs) removal, the recovery yield and the dynamic capacity are after that, determined with the selected conditions using 5mL-scale columns. Optimization and scalability have been finally performed with larger columns and bed heights compatible with industrial constraints.
Efficiency (recovery yield and purity improvement) of each step of the developed purification protocol has been evaluated by different analytical tools, including SDS-PAGE (Fig.6), from the extraction step to the final formulation by ultrafiltration.
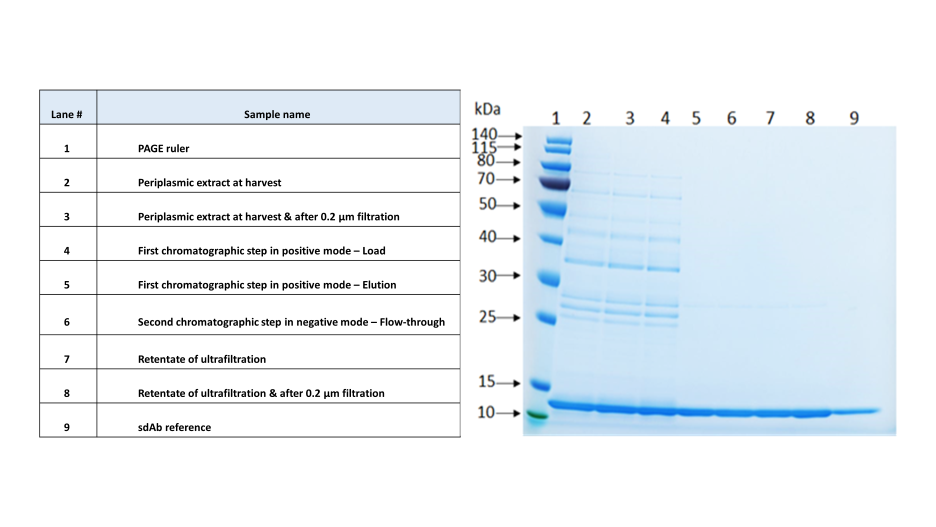
Fig.6 – Monitoring of the purification protocol of a sdAb by SDS-PAGE
Development of the In Process (IPC) and Quality Controls (QC)
IPC
For each step of the manufacturing process, evaluation of the expression yield (by SDS-PAGE and WB), recovery yield (by UV and/or µBCA assay), purity improvement (by SDS-PAGE and/or RP-HPLC), protein integrity (by RP-HPLC) and protein aggregation (by SE-HPLC) are implemented (Fig.7). HCP and endotoxins removal are also evaluated.
QC
The purified and filtrated sdAb has been characterized using a large range of analytical methods:
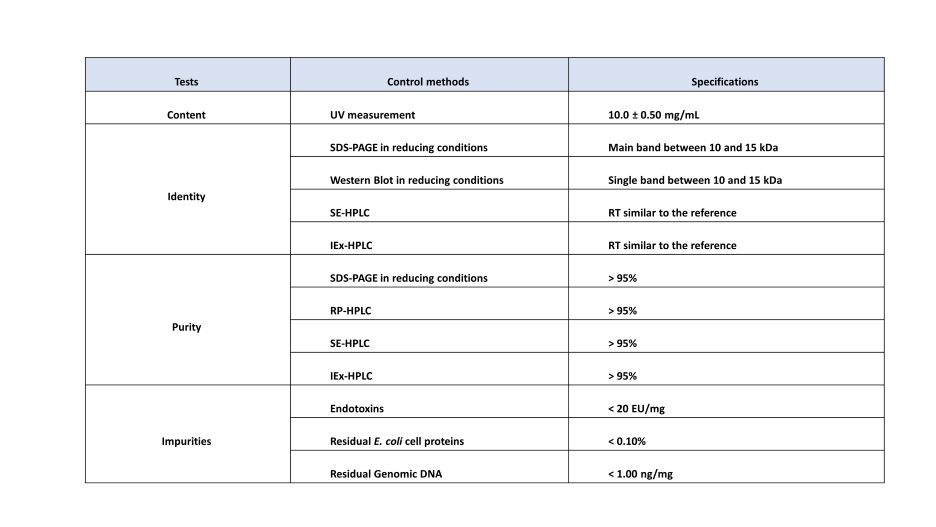
Purity analysis by SDS-PAGE (Fig.8):
The equivalent of 20 µg of sdAb is loaded in lane 2,; the sample is diluted twice and loaded in the following well, up to 78 ng is loaded in lane 10. After densitometry analysis, the purity was evaluated at > 99.5%.
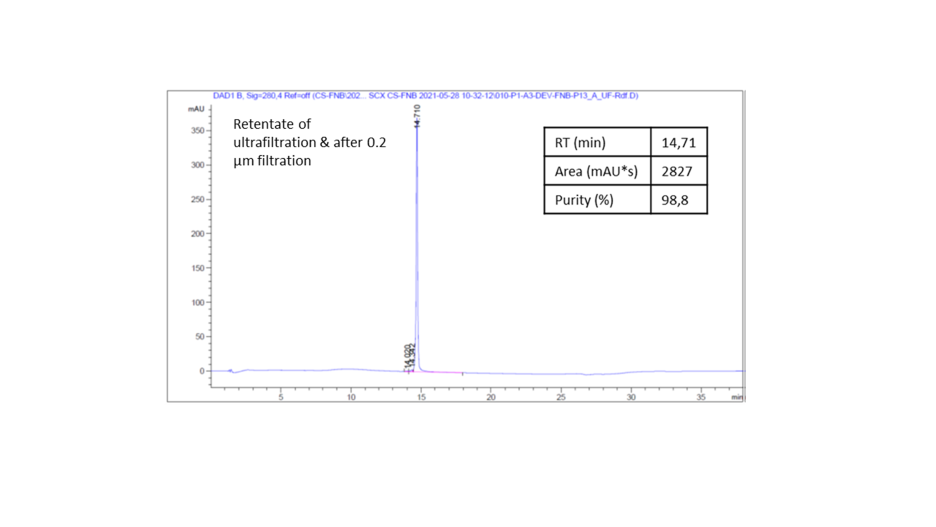
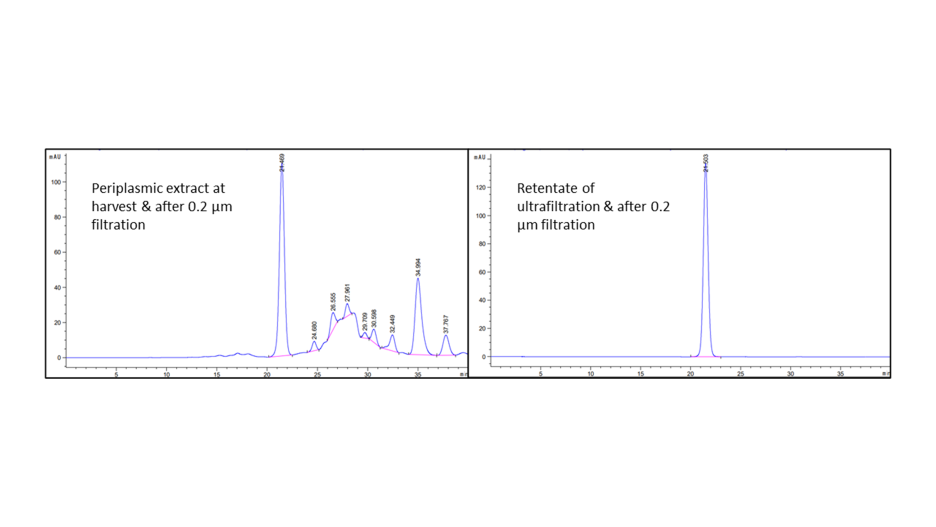
Purity analysis by IEx-HPLC (Fig.9):
The aim of this IEX-HPLC is to analyze the purity level at the completion of the purification process and the presence (or not) of isoforms or host cell proteins (HCP). This test is also used as identity test. After peak speak integration, the purity was evaluated at 98.8%.