FectoPRO is a transfection reagent specifically optimized for transient protein and antibody expression in CHO and HEK293 cells.

When expressing recombinant proteins, in any given cellular model, one goal might be to harvest the recombinant product from the culture media. This avoids the necessity to lyse the producing cells to obtain the product, hence reducing the risk of protein contamination through the subsequent purification steps. You may also need your recombinant protein to be glycosylated, which require to use cells from upper organisms and to target the protein to the secretory pathway.
For both reasons, and possibly others, you need to use a system to address your extending protein to specific locations in the cell. Actually, Nature devised such systems that are used to sort protein in the cellular compartments. One of them relies on Signal Peptides recognized by the Signal Recognition Particles (SRPs).
There are several secretory pathways relying on N-terminal signal sequence that have evolved in various organisms1. In this chapter we will focus on SRP-dependent pathway, as this one prevails in eukaryotes, but others can be named here, such as the sec-dependent or tat-dependent pathways that are prevalent in bacteria. Many of these pathways however co-exist within the same cells and organisms.
Signal Recognition particles are RNA-protein complexes with a conserved function but an evolving structure from Bacteria to Mammals. Simples bacterial SRPs are formed of the 4.5S RNA complexed with Ffh2, Ffh7 and Ffh8 proteins. Mammalian SRPs consists of the 7SL RNA and six proteins (SRP54, SRP19, SRP68, SRP72, SRP14 and SRP9). However, their structural evolution their biological function remains closely related: to bind to a translating ribosome and its nascent peptidic chain and to recognize a signal peptide on the peptidic chain emerging from the ribosome (figure 1). Upon recognition of such signal peptides, the translating ribosome is docked to a protein conducting channel2. In bacteria this channel is present on the plasma membrane, in Mammals it is located on the reticulum membrane.
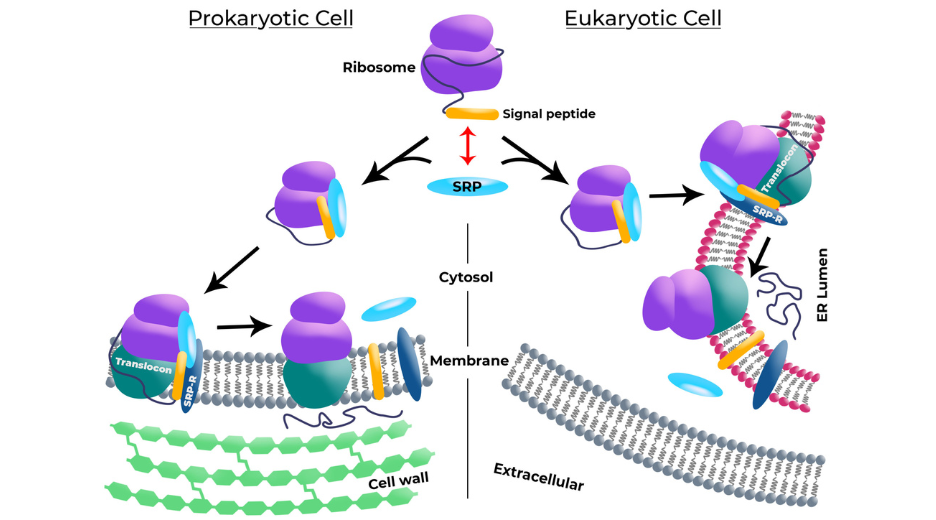
Figure 1: Signal peptides mechanism in Prokaryotic Cell and Eukariotyc Cell.
Despite the diversity of existing the secretory pathways, signal peptides have conserved common structural features across the species. Three regions, termed N-, H- and C-region, have been studied and characterized.
N-region is a short (1-5 residues) positive-charged domain. Its basic amino acids (Arg or Lys) are preferentially encoded by high-translational-rate codons. The charges are involved in the interaction with SRPs as well with the phosphate of the lipid layer. They impact the orientation of the SP in the membrane in order to maximize the SP availability to SPases (see below).
H-region is an hydrophobic helix (7-15 residues) that will be anchored in the membrane. This region is very specific of its species which make it predictable (75-90%) within a specific species3. Leucins are predominant in such domain and 3-5 residues of leucin seem to positively affect secretory efficiency.
C-region is a beta-sheet folded uncharged region (3-7 residues) that will serve as substrate for the SPase. SPases are the enzymes that will cleave the SP and release the protein in the secretory pathway. A motif V/A-X-A is highly conserved in eukaryotes just before the cleavage site, while eukaryotes SPases seem to tolerate a wider X-X-A motif.
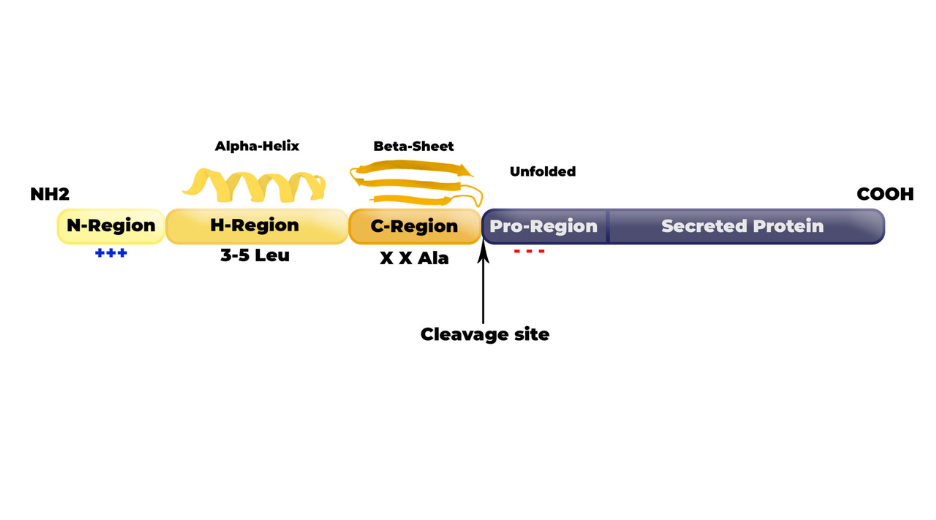 Figure 2: Signal peptide structure.
Figure 2: Signal peptide structure.
Beyond the cleavage site, the Pro-region (i.e the 1-6 residues starting sequence of the secreted protein) will also affect the SPase activity. Preferably, amino-acids in this Pro-region should be small and flexible as well as neutral or acidic to avoid steric hindrance or unwanted folding that would decrease the SPase efficiency.
Albeit appearing very similar, signal peptides and secretory pathways seems to be very finely regulated. Many studies report drastic changes in secretion when only one amino acid of the SP is modified (see for review1). This is also true when one considers the Pro-region that is strictly dependent on the protein of interest.
For example, Haryadi et al.4 have compared the expression rate of various antibodies by CHO according to the nature of the SPs fused to the heavy chain. As shown in Figure 3, expression rates can be very variable for a given antibody depending on the Signal peptide (H1 to H8). Furthermore, although the SP H7 seems to be the most efficient four 4 tested antibodies, it appears to be the less efficient one for the 5th antibody (Herceptin). This suggests that the interplay between the secreted protein sequence and the SP placed upstream plays crucial role in the expression/secretion rate.
Similar works conducted on prokaryotes led to the conclusion that no universal SP exists that promotes the most efficient secretion for any recombinant protein, in any chosen model. Instead, an optimal SP has to identified for each protein in each expression system by a screening strategy5
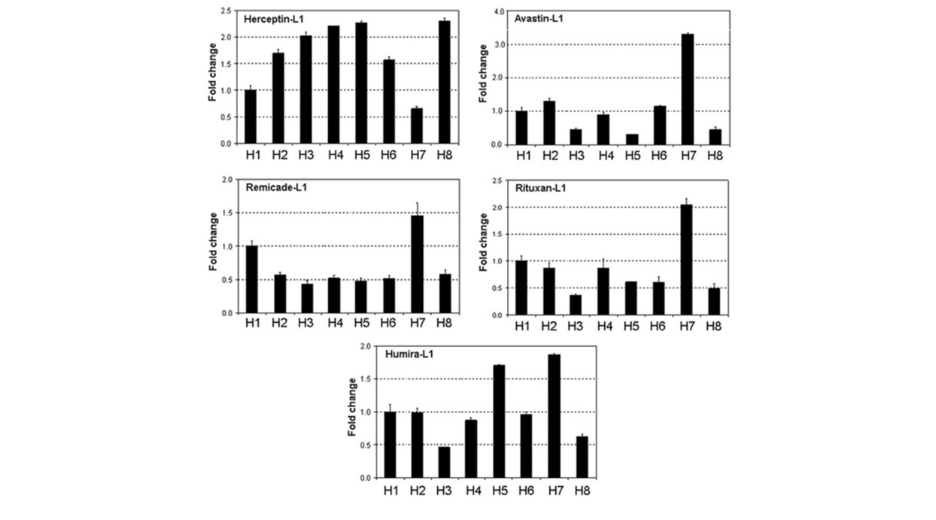
Figure 3: Expression rate of Herpcetion, Avastin, Remicade, Rituxan and Humira by CHO. Expression is variable for a given antibody depending of signal peptide (H1, H2, H3, H4, H5, H6, H7 and H8). Haryadi, R. et al. Optimization of Heavy Chain and Light Chain Signal Peptides for High Level Expression of Therapeutic Antibodies in CHO Cells.
Tip 1: Pick SPs belonging to the species you wish to express your protein in. You will increase your chance that the SP will be properly handled by the secretory pathway of your expressing cells. There are many ‘commonly used’ SP that people use in most-used mammal cell lines (CHO, HEK-293 or Hela), some of them you will fin on our public library on D-Zyvec (e.g. Azurocidin or Serum-albumin). You can also find many described SPs in specific databases such as the Signal Peptide Website.
Tip 2: Include at least 2 SPs in your first set of constructs. When working for the first time with a given protein, you will decrease the risk of randomly picking the worst SP possible for your protein. If you can, try 3 to 5 SPs simultaneously to identify the best candidate that can be then further modified to improve expression rate if necessary. Of course, this mean to multiply the DNA construct to do so, but remember that our e-Zyvec’s team just happen to be specialized in serial vector building. We can help you to save a lot of time at the beginning of your projects.