Restriction Enzymes and Molecular Cloning
Slightly after the discovery and understanding of plasmids (during the fifties) scientists discovered and studied proteins able to recognize and cut double strand DNA molecules (during the seventies). These catalytic proteins are known as ‘restriction enzymes’.
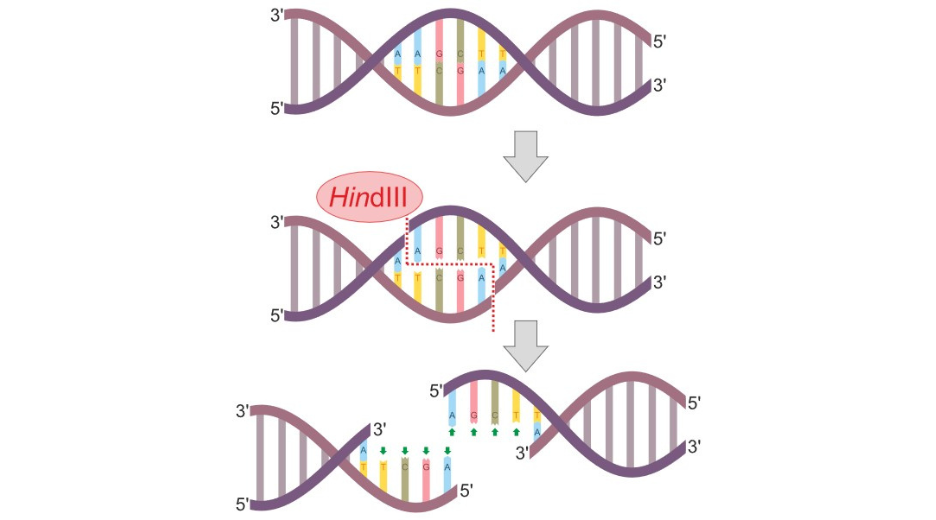
The natural function of restriction enzymes is to act as a defense mechanism against viruses infecting bacteria. Indeed, bacteria like any other cell can be infected with viruses, called bacteriophages, that will inject their own DNA in the bacteria in order to propagate themselves. Restriction enzymes selectively appeared and evolved so the bacteria could cut, or ‘digest’ the viral DNA, thus preventing the viral propagation. Basically, a restriction enzyme is able to recognize a short DNA sequence (4 to 8 base-pairs long on average) and to actively incise both DNA strand, either within the recognition site or at some distance, depending on the enzyme. The digestion thus produced DNA fragment with either ‘blunt’ or ‘cohesive’ ends of defined sequence.
Evolution has produced a wide variety of restriction enzymes differing by their recognition sequence (also called restriction sites) and mechanism of actions. More than 600 have been isolated and engineered to be used by biologists to digest DNA molecules at specific site.
So, by the end of the seventies, biologists had auto-replicative plasmids in one hand and restriction enzymes in the other. It did not take them long to start using the later to introduce DNA sequences of interest in plasmids, thus creating hybrid DNA molecules, amplifiable by bacteria. Indeed, if one could cut two DNA molecules, one being a plasmid, in a similar sequence specific way, DNA fragments could be assembled on the base-pair complementarity basis. To make this work in an easy way, the system needed a couple more elements.
First, biologists needed another type of enzyme that would be able to stitch the complementary DNA fragments together. These enzymes naturally exist in every biological organism where they are involved in DNA repairing or DNA replication. They are called DNA ligases. Ligases from bacteriophages, such as T4 ligase, have been engineered to be efficiently used to seal DNA fragments together. They are now widely used in molecular cloning activity.
The second requirement to perform efficient cloning in plasmids is to work with restriction enzymes which recognition site is only occurring once in the plasmid sequence. Furthermore, this occurrence must not be located within any functional feature of the plasmid (e.g. Origin of replication or Marker of selection) so the functional integrity of the vector remains intact. Thus, to render easier the use of plasmids as vectors for amplifying or expressing reconstituted genes, scientists have developed the concept of multi cloning site (MCS).
An MCS, also called polylinker, is a short segment of DNA that contains several restriction sites (up to 20) that are unique in the vector. The presence of an MCS in a cloning vector or an expression vector (downstream a promoter), ensure the ability to ‘insert’ the gene of interest at a specified location within the vector. Having several restriction sites in the MCS increases the chance that at least one of them will not be randomly present in the sequence of interest (SOI) to be inserted in the plasmid. It may also help to orientate the SOI in the vector by selecting two different sites to be used at each extremity of the DNA fragments to fuse.
From the seventies until now, the formidable collection of engineered restriction enzymes, as well as the exponential development of commercial vectors based on plasmids, have been allowing the biologists to undertake molecular cloning quite efficiently for their genetic engineering projects. However, as knowledge in biology increases, researchers are more and more confronted to complex biological questions that requires multiparametric assays to be set up. Classical molecular cloning as described above, although designed to be very versatile, is challenged by the emergence of new experimental requirements, and starts to show its own limitations.
Here are some of these limitations:
- Plasmid derived vectors are by definition some single purpose molecular tools. Thus, when several applications a required to complete a project, molecular cloning must be performed as many times as required, by changing (purchasing) the vector.
- Vectors content being different, the MCSs also vary, and are not always fully compatible between each other or with the SOIs
- Although tens of thousands of vectors have been reported in the literature (over 183k publications on pubmed, 65k vectors in Addgene’s repository) each new project requires to find the appropriate tool amongst this vast library, or, more often, to recreate a new one that will fit the specific requirements. In other words, standardization of vectors only partially address the needs.
- The use of MCS in plasmids is contradictory with the growing needs of working with 2 or more SOIs simultaneously. Vectors with multiple MCS are neither easy to create nor easy to use.
So, 40 years after the dawn of molecular cloning, and to address the extraordinary vivacity of the field, we believe it is already time to revisit the ‘old ways’ and contribute to the evolution of genetic engineering.
