FectoPRO is a transfection reagent specifically optimized for transient protein and antibody expression in CHO and HEK293 cells.

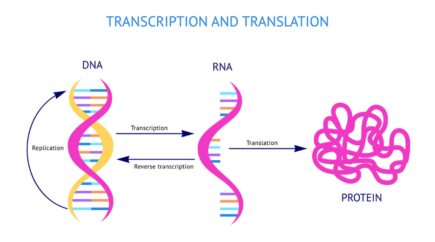
Protein biosynthesis is the result of the RNA to Protein translation process.
Messenger RNA (mRNA), transcribed from DNA, will be processed by the ribosomal machinery to produce the corresponding protein. Using the genetic code, ribosomes will recruit transfer RNA (tRNA) matching the read codons. Amino-acids brought together within the ribosome will be linked with a peptidic bond and become part of the future polypeptide chain encoded by the mRNA. The nucleotide sequence encoding a polypeptide from its start codon to its stop codon, is referred as
CoDing Sequence(CDS) pending on the biological system (eukaryote or prokaryote) translation mechanistic is slightly different. This needs to be considered when designing gene expression vectors.
By definition prokaryotes do not possess a subcellular compartment isolating the chromosomic DNA from the cytosol. Therefore, DNA transcription to mRNA and mRNA translation to peptide chains can occur simultaneously. Indeed, protein transl
ation is active on mRNA that are still being transcribed from the genomic DNA.
Another specificity of the eukaryote genes is that most CDS are grouped in polycistronic operons. This mean several CDS are transcribed in a single mRNA molecule, each of them being preceded by a ribosome binding site (RBS), which is the sequence directly upstream the start codon.
Translation in bacteria starts with the recognition of a consensus sequence (5′-AGGAGG-3′) in the RBS, known as the Shine-Dalgarno (SD) sequence1, by the small subunit of (30S) of the ribosome. The recognition is based on base-pairing complementarity, the complementary sequence (anti-SD) CCUCCU being present in the 3’ end of the rRNA part (16S) of the subunit. The SD sequence is optimally located 6 nucleotides upstream of the start codon2.
Involvement of three proteins known as initiation factors IF1, IF2 and IF3 completes the initiation complex. The conformation of this multimolecular complex brings the AUG start codon of the mRNA in front of the P site of the 30S subunit. IF2 is then recruiting the initiator tRNA to the complex, which in turn allows the recruitment of the large subunit (50S) to form the complete and active 70S ribosome3.4. In bacteria the initiator tRNA is carrying a N-formylmethionine that will be the first amino acid in the nascent peptide and will be removed during the elongation step.
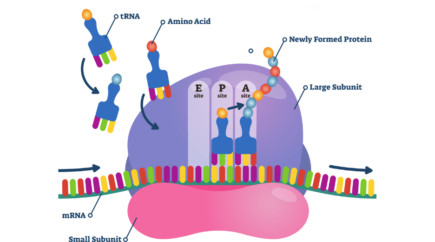
Starting from the initiating amino acid (a N-formylmethionine), the elongation of the polypeptide chain involves addition of amino acids to the carboxyl end of the growing chain. The fully assembled ribosome displays three ‘pocket’ sites where tRNA can insert so to bind to the mRNA presented by the small subunit (30S) in a complementary way. These binding sites are designated, from 5’ to 3’, E, P and A. The E-site (exit) accommodate a free tRNA; the P-site accommodate a peptidyl-tRNA (a tRNA bound to the poly-peptide chain) and is connected to the polypeptide exit tunnel; and the A-site accommodate an aminoacyl-tRNA or termination release factors.
With the initating tRNA located in the P site, a conformational change ‘opens’ the A site, so the ribosome can harbor a new tRNA. The next tRNA to come will be selected based on its complementarity to the mRNA on the three nucleotides downstream the AUG codon, that is the downstream codon. Amino acids brought together in the P and A sites are linked together with a peptidyl bound to extend the growing protein chain.
The extending chain is this way transferred in bulk on the upcoming tRNA (in site A) and in the process the ribosome is shifting along the mRNA so each tRNA switch binding site. The released tRNA that was in P site moves to the E site, pushing out the previous free tRNA if there was one, and the upcoming tRNA, now carrying the peptide chain moves to the P sites. The A site being free again, a new tRNA can be harbored to match the next codon. The nascent protein exits the ribosome through the polypeptide exit tunnel in the large subunit (5
0S).
Ribosome carries on reading the mRNA that way until it meets a stop codon.
In the genetic code, three codons are not corresponding to amino acids but instead act as translation termination signal. These termination codons are UAG (“amber”), UAA (“ochre”) and UGA (“opal” or “umber”).
Termination occurs when one of the three termination codons moves into the A site. These codons are not recognized by any tRNAs. Instead, they are recognized by proteins called release factors, namely RF1 (recognizing the UAA and UAG stop codons) or RF2 (recognizing the UAA and UGA stop codons). These factors trigger the release of the newly synthesized protein from the ribosome.
Experimental data indicate that UAG codon is universally suboptimal in bacteria. The ochre stop codon UAA is likely to be the preferred stop codon for bacteria with geno
mic low GC content while the opal UGA is the preferred stop codon for high GC content bacteria. The optimization of stop codon usage may therefore be useful in genome engineering or gene expression optimization applications5.
At last the phenomenon of stop-codon readthrough has been described in many cells including bacteria6. Deopending on the stop codon, the strain and the condition of culture, a small percentage of translation (usually <10%) is not terminated properly. Nucleotide sequence downstream is then translated creating a bigger protein, which function (or disfunction) is difficult to predict. One possibility to reduce stop codon readthrough is to add one or two other stop codons, possibly of the other types, after the first one.
Eukaryote defining features is the presence of a nuclei in the cell that will separate the genomic DNA from the rest of the cell. Compartmentation has permitted to cells to complexify their genome and gene structure. Now that translation is separated in time from transcription, mRNA can be produced as an immature form that can be further processed (mRNA splicing) to obtain a mature mRNA to be exported to the cytosol for translation.
This new step gives the cell more option to regulate its protein content since an initial transcript can give rise to alternate versions of a protein by alternate splicing. Maturation of the mRNA also includes the polyadenylation of the its 3’end.
Once exported to the cytosol through nuclear pores, matured mRNAs are translated by ribosomes in a very similar way to what happens in prokaryotes. Below are the differences to take into account when designing expression vectors.
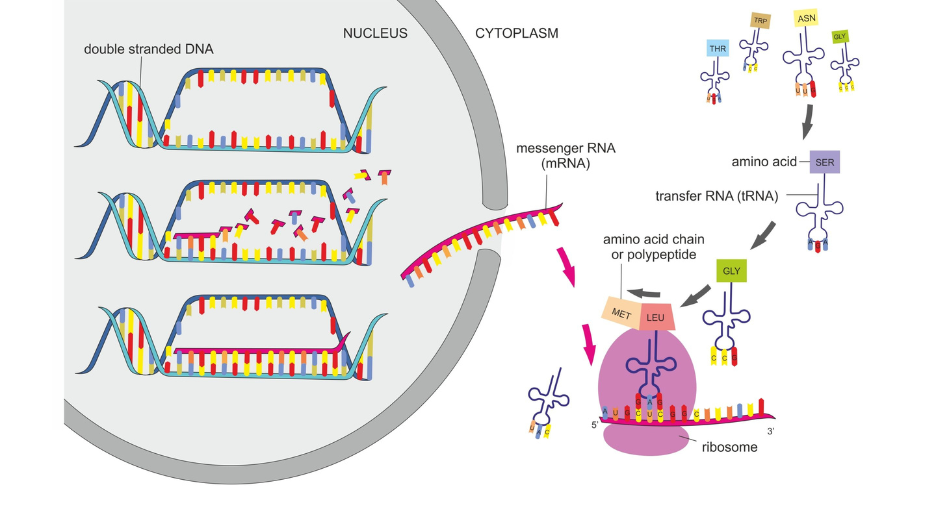
There are no RBS in eukaryotes to drive the entry of the ribosome next to the start codon AUG. Instead the initiation complex formed of several proteins (eIF2, eIF3 and eIF4), the initiator tRNA and the small unit of the ribosome (40S in eukaryotes) assembles à the 5’Cap of the mRNA. In most of the natural eukaryotic mRNA, the start codon AUG is not in the vicinity of the assembly site. Thus, the initiation complex will move along the mRNA molecule toward its 3′-end, in a process known as ‘scanning’, to reach the start codon.
The nucleotide sequence that’s is preceding the AUG is defined as 5’ UnTranslated Region (5’UTR)to distinguish it from the CDS. 5’UTRs are important for the regulation of translation of a transcript through various mechanisms. Depending on the purpose of your vector, you may want to consider including the 5’UTR (and the 3’UTR) to your expression cassette, to ensure an expression as close as possible of the physiological conditions.
The AUG starting the CDS is comprised into a larger consensus sequence 5’-(gcc)gccRccAUGG-3’ named Kozak Sequence. When reaching the AUG, base-pairing complementarity with the tRNA in the initiation complex triggers the complex dissociation and allows the large subunit (60S) to assemble with the small one. In that context, it has been postulated that the Kozak sequence also interacts with the 40S one, inducing some conformational changes presumably helping the transition to a complete and functional ribosome7.
Since the description of the Kozak sequence for vertebrates, the concept of the consensus or optimal translation initiation sequence has been refined by several researchers. It was shown that, presumably as a reflect of the evolution, the upstream sequence of the AUG codon varies according to the taxonomic group8 and to the GC content of the 5’UTR9. Therefore, when designing a gene (protein) expression vector, one may consider selecting the fitter consensus of Kozak sequence according to its system of expression.
At last, a 5’Cap independent initiation of translation has been thoroughly described, relying on Internal ribosome entry site (IRES). This is detailed here (button)
There is no major difference in the translation mechanistic between eukaryotes and prokaryotes. However, the structure of the molecules involved in translation must display subtle differences since ‘self-cleaving’ 2A peptides are efficient in eukaryotic cells but not in prokaryotic ones as detailed in the 2A page (button)
This step is identical to what occurs in prokaryotes. When meeting a stop codon the ribosome will release the peptide chain and disassemble itself.

FectoPRO is a transfection reagent specifically optimized for transient protein and antibody expression in CHO and HEK293 cells.
jetOPTIMUS® is an innovative cationic nanotechnology developed to improve DNA transfection efficiency in hard-to-transfect cells.
jetPRIME® is a versatile DNA and siRNA transfection reagent that ensures high DNA transfection efficiency and excellent gene silencing.

Polyplus provides an integrated service for design and production of tailor-made DNA plasmid through our unique and Innovative assembly technology.

Polyplus offers a comprehensive range of plasmid manufacturing services from DNA plasmid design to plasmid DNA manufacturing and analytical testing.