FectoPRO is a transfection reagent specifically optimized for transient protein and antibody expression in CHO and HEK293 cells.

Engineering protein often relies on fusing different protein domains to combine their biological functionalities. However, proteins are three dimensional objects, and the juxtaposition of protein domains has to be considered not solely in a linear way, but also in space. In other words, when fusing different domains, one has to envisage how these domains will interact. Do they need to be close or spaced in order to interact? What if we need them to not interact at all?
Simply put, this gets back to evaluating two main criteria: size and flexibility of the linker. It is impossible to edict cast in stone rule as to what linker must be used in which condition. Ideally several design should be tested and compared; would it be just to demonstrate that there is no bias in the observation due to the nature of the linker. But to help your thinking and choices here are some tips and advice:
There is no absolute rule stating that a linker is essential for a given protein fusion. It will be mostly dependent on the final conformation of each of the protein domain.
Simply bear in mind that although ‘linearly’ synthesized by the sequential addition of amino-acids, polypeptide may also go through some complex folding process. By combining two highly structured domains close to each other, you may risk impairing the folding of each or both domains. If the protein structure is known and shows unfolded region next to the fusion, this risk is mitigated. If the structures of the domains are not known, you may wish to insert a small flexible linker, just in case.
Such linker can be used to give some room to each domain, to favorize protein folding, to limit unwanted interferences between the domains or to remove a domain from the membrane vicinity in the case one of the domains is a transmembrane one.
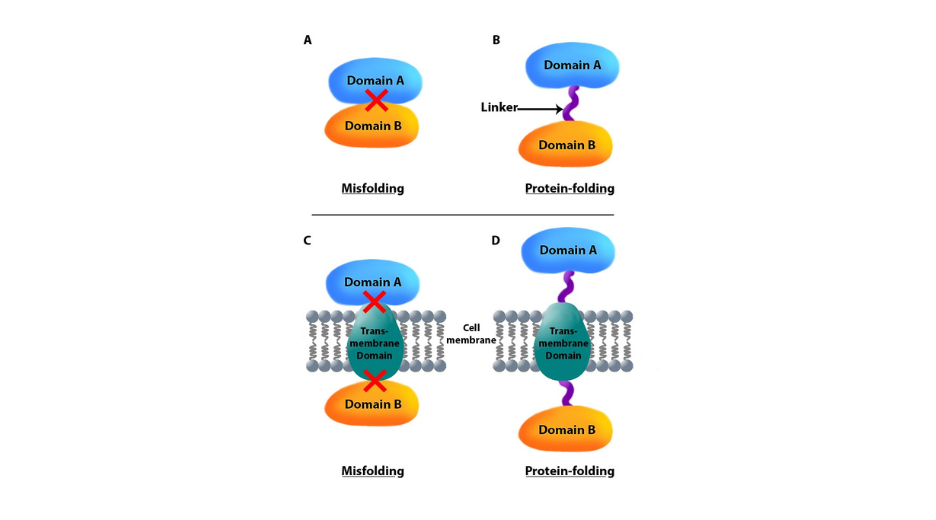
Figure 1: A linker may help proper folding.
In such cases we recommend using flexible spacers of 3-5 amino acids length. Commonly used linker are stretches of small polar or non-polar amino acids, such as Glycin (Gly/G) and Serine (Ser/S). One of the most used linker is (GGGGS)n, where n can be adjusted to create longer linkers1.
There may be cases where short linkers are not sufficient to ensure a proper expression or functionality of both domains. Indeed you may need each domain to get enough freedom of movement to interact with their targets/partners, or even to properly interacts with each others.
In longer linker, Glutamate (Glu/E) or Lysine (Lys/K) can also be used to increase solubility, as exemplified in the table below:
| Examples of flexible peptide linkers |
| (GGGGS)n 2 |
| KESGSVSSEQLAQFRSLD 3 |
| EGKSSGSGSESKS 3 |
| SSGNSNANSRGPSFSSGLVPLSLRGSH 4 |
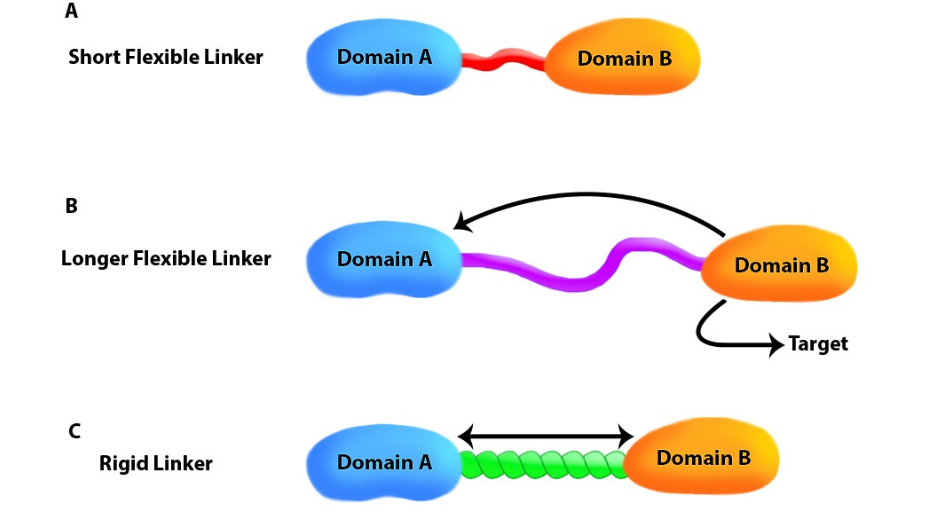
Figure 2: The right link for the right biological function. A) Each domain can be active individually. B) Lenght and flexibility to favor interaction with third peptide. C) Avoid domain interaction (rigid).
There may be some cases where the purpose of the linker is also to make sure that the two domains to be fused remain well apart from each other. In such case, flexible linkers, which main characteristic is to be unfolded, may not be sufficient. Indeed, a long flexible linker will not naturally extend to its maximal length, at least not all the time, but will rather coil on itself, because of its flexibility, allowing the two domains to get close to each other. If you wish to keep your fused domains well separated, you may prefer to use folded linkers that would act as rigid spacer.
Several such domains have been assayed to compare the flexibility conferred to the chimeric proteins2: for instance Zn-a2-glycoprotein (ZAG; PDB code: 1ZAG) that is a relatively elongated and rigid structure, or b2-microgloblin (b2m; PDB code: 1LDS) and ubiquitin (Ub; PDB code: 1UBQ) that are small globular proteins). However, these domains will bring their own immunogenicity as well as possibly some functional features, to the fusion. Furthermore, they must be used as a whole to keep their structure, thus there is no possible modulation of the size of the linker, which can be quite large compared to the domains of interest.
Another possibility is to use alpha-helix based linker. Helix structure will provide the wanted rigidity of the spacer. The size of the spacer can then be modulated by changing the size of the helix and/or the number of repeats2,5.
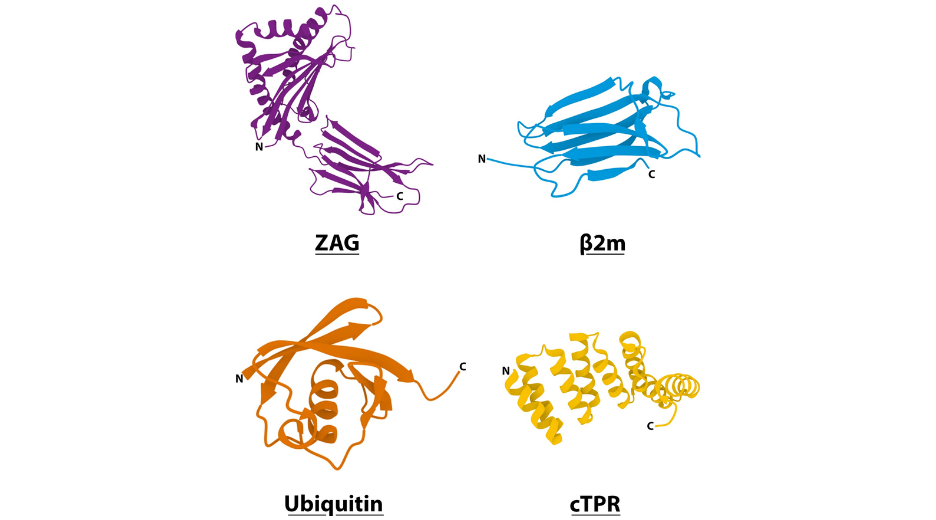
Figure 3: 3D visualisation of domains used in structured peptide linkers (pdb codes: ZAG (1ZAG), β2m (1LDS), ubiquitin (1UBQ), cTPR (2FO7)).
5.1) Keep in mind that, depending on the molecular context, linkers can either 1/ help or impair the proper folding or localization of the fusion proteins, 2/ push apart (rigidity effect) or bring together (flexibility effect) the fused domains. This is particularly important to assess when you wish the two domains to interact, like with FRET/BRET experiments6.
5.2)Enzymatically cleavable peptides exist (e.g. Furin, TEV sensible peptides)that can be used as linker in fusion proteins. They are usually unstructured flexible linkers. Depending on your experimental set up, you may find interesting to be able, in a second step, to separate the fused domains in a controlled way.
5.3) If the relative position of fused domains is not essential for the desired functions, or if its effect is simply not known, we recommend testing the fusions both ways in your preliminary experiments. That means fusing Domain A to B (A-linker-B) and B to A (B-Linker A), to compare the biological features of both fusions. Of course, due to domain conformation (if the structure is known) or subcellular localization, such permutation may not make sense at all. But if you reached such conclusion, it means you have asked yourself the right questions!
5.4) At last, always compare at least two linkers when first fusing two domains. You can also go with no linker at all as another comparison point. So, if you multiply the linkers (or not) in terms of structure or length, plus the relative position of your domains, you may realize that your pilot experiment will soon require 4 to 12 different constructs. Sounds complicated to pull off? Don’t worry, combinatorial assembly is exactly what e-Zyvec is good at. In the end, a good rationale in your choices of fusions will always make your results look more convincing and will provide you good controls or back-ups.