Polyplus provides an integrated service for design and production of tailor-made DNA plasmid through our unique and Innovative assembly technology.

Today we will be telling you about the largest plasmid we have been challenged to assemble by a customer, so far. Let’s call it Viktor 23K. There is no point to build large plasmids just for the sake of their size, quite the opposite actually. What matters is to integrate as many genetic features as possible within a single molecule. So, what was Viktor about?
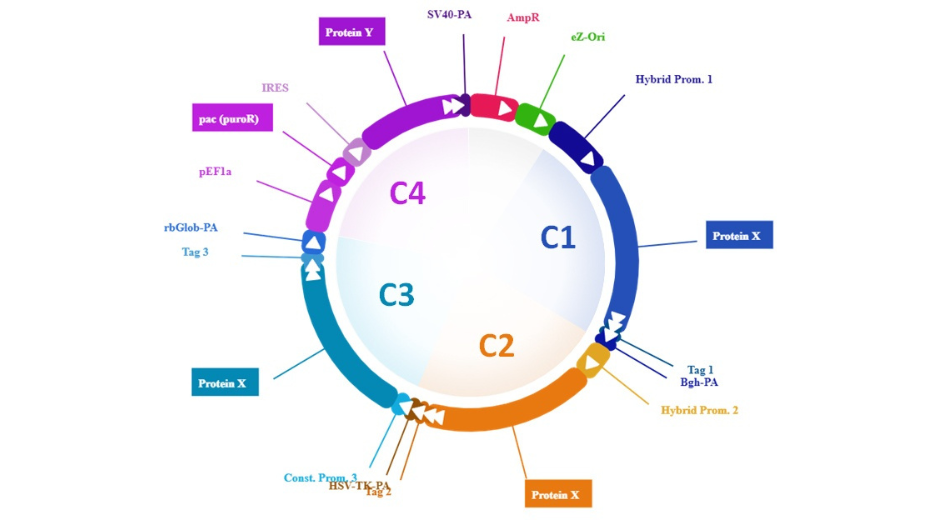
The aim was to overexpress a single protein (Protein X) as much as possible, using not one single, but three different expression cassettes (C1, C2 and C3). Each cassette was driven by a distinctive strong promoter. So, not only the vector would trigger an improved protein expression, but this expression was expected to be somewhat less dependent on environment changes, thanks to the variety of the selected promoters. Actually, individual cassette expression could even be assessed thanks to the presence of three different peptide Tags (Tag1, Tag2, and Tag3) fused with protein X in each cassette.
Viktor also contains a fourth cassette encoding two more genes. The first is an eukaryotic selection marker (for puromycine selection). That’s because our customer also wanted to select stable transfected cells. Therefore, we also packed a complete recombination system (unannotated) so Viktor would integrate in full in the target cells. The second is an enzyme (Protein Y) that is expected to act on Protein X to improve its biological function. Now these two genes are encoded within a single transcript with the use of an IRES. This helped save a little weight and was possible because it was known that Protein Y could be effective even at a low expression rate. This is rational design.
Proofreading Viktor:
Being a big guy as he is, we could not assemble Viktor in one go as we usually do with regular plasmid. Instead, we first assembled smaller blocks (like cassettes) from various parts, and then got the block together. Anybody who built large Lego models recently will exactly grasp the concept.
But how to check eventually that Viktor was exactly what we promised it would be? That is, how to assess the sequence in a fast and affordable way? Luckily, we have recently developed our own NGS analysis method to proofread our constructs. Raw data obtained from an Illumina platform are automatically processed to compare the NGS results with the predictive sequence. This takes only a couple of minutes per plasmid!
However, Viktor was not the kind to make it easy to us. As you may remember, there are three cassettes containing each the same coding sequence for protein X. Long repeated sequences as these can make things tricky when it comes to NGS. This is why we coupled our routine NGS analysis with a restriction print analysis selecting restriction sites in order to make sure that all three cassettes actually contained the Protein X sequence in full.
Cutting a long story short: Viktor turned out not to be absolutely what we expected. We found one single mutation, fortunately one without functional incidence. Viktor was only 99.996% what we predicted… We are sure we can do better than that!