The DNA molecule is composed of two intertwined polymeric chain of deoxyribonucleotides (the building blocks of DNA), forming the so-called DNA double helix. Each DNA chain, or strand, is complementary to the other according to its nucleotides sequence, each G matching C and each A matching a T on the opposite strand and vice-versa.
DNA molecules can be copied, or replicated, by an enzymatic machinery called DNA polymerase complexes. But the way DNA polymerase work is by reading single strand DNA molecule and building up a new complementary DNA strand by linking free deoxyribonucleotides together, one by one as it reads the existing strand. This implies that the existing DNA strands of the initial DNA molecule have to be separated for the DNA polymerase to be able to copy them. Separating the strands of a DNA double helix is called DNA denaturation or DNA melting.
Plasmids are circular DNA molecules that can be present in multiple copies in bacteria. One can figure them as two intertwined rubber-bands (each band representing a DNA strand). To benefit from the plasmidic ‘bonus’ genes, the bacteria has to be able to duplicate the plasmid in order maintain its pool after passing on some copies to other bacteria (during conjugation) or after it divided into two new cells (cell division). Therefore, plasmids have to come with a way for them to be melted (opened) somewhere, so DNA polymerases can start reading each DNA strand and duplicate them.
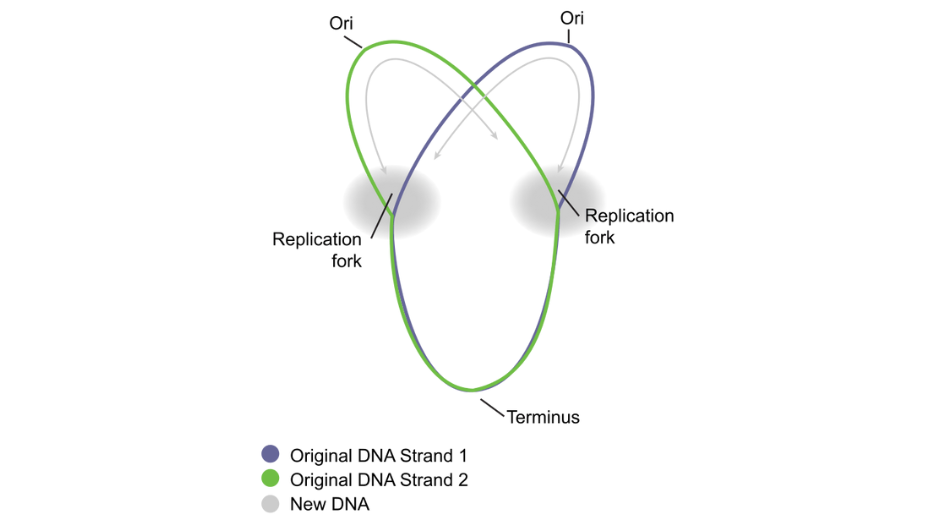
Illustraion of plasmid replication showing the two replication fork. Original, David Tribe, derivative, Rehua. Creative Commons Attribution-Share Alike 3.
Such a region exists in every plasmid and is called ‘Origin of Replication’. There are several different sequences described to function as Origin of Replication, differing from one plasmid ‘strain’ to another, according to its strength (numbers of copies induced within a single bacteria) or according to the strain of bacteria the plasmid will be amplified in. Some of the best described are ColE1, pMB1 or p15A. They share a general mechanism relying on the transcription of non coding RNA that will form secondary structure to maintain the plasmid DNA strands separated. These RNA-DNA interactions allows further recruitments of enzymes (RNAse H and DNA polymerase) and the initiation of new DNA synthesis. Eventually the full plasmid will be duplicated by the enzymatic machinery of the host bacteria cell.
Origins of replication are therefore paramount for the plasmid/vector amplification in bacteria and will determine the yield of production of DNA in bacterial culture.